Written by
Seewoo
on
on
Constructing Hospital Score Using PCA
1. EDA
reading csv file
merging plots
hpdata <- inner_join(table_1, table_2, by='hpid')selecting variables
hpdata <- hpdata %>%
select(dutyName.x, starts_with('h'), starts_with('mk'))%>%
select(-hv1, -hv12, -hvidate)
glimpse(hpdata)## Rows: 313
## Columns: 36
## $ dutyName.x <chr> "의료법인강릉동인병원", "강릉아산병원", "강원도삼척의료원", "강원도속초의료원", "의료법인보광의...
## $ hpid <chr> "A2200005", "A2200008", "A2200007", "A2200012", "A220004...
## $ hv10 <chr> "0", "Y", "0", "0", "0", "0", "0", "Y", "Y", "Y", "0", "...
## $ hv11 <chr> "0", "Y", "Y", "0", "0", "0", "0", "Y", "Y", "Y", "0", "...
## $ hv2 <dbl> 6, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0,...
## $ hv3 <dbl> 0, 1, 0, 0, 0, 0, 0, 2, 0, 0, 0, 0, 0, 0, 5, 10, 4, 0, 0...
## $ hv4 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hv5 <chr> "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "...
## $ hv6 <dbl> 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hv7 <chr> "Y", "Y", "0", "0", "0", "0", "0", "Y", "Y", "Y", "0", "...
## $ hv8 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hv9 <dbl> 0, 0, 0, 0, 0, 0, 0, 4, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hvamyn <chr> "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "...
## $ hvangioayn <chr> "N", "N", "N", "N", "N", "N", "N", "N", "N", "N", "N", "...
## $ hvcc <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hvccc <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hvctayn <chr> "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "...
## $ hvec <dbl> 17, 14, 18, 19, 8, 4, 8, 15, 14, 16, 9, 10, 10, 10, 13, ...
## $ hvgc <dbl> 64, 62, 15, 59, 37, 148, 90, 91, 143, 42, 73, 19, 30, 10...
## $ hvicc <dbl> 1, 0, 1, 3, 6, 3, 0, 0, 0, 0, 11, 1, 0, 6, 0, 0, 0, 4, 4...
## $ hvmriayn <chr> "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "...
## $ hvncc <dbl> 0, 6, 0, 0, 0, 0, 0, 17, 10, 0, 0, 0, 0, 3, 13, 4, -1, 0...
## $ hvoc <dbl> 5, 13, 2, 3, 4, 3, 6, 15, 11, 9, 2, 2, 0, 8, 10, 2, 13, ...
## $ hvventiayn <chr> "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "...
## $ mkioskty25 <chr> "Y", "Y", "N", "Y", "Y", "N", "Y", "Y", "Y", "Y", "Y", "...
## $ mkioskty1 <chr> "N", "Y", "Y", "N", "N", "N", "N", "Y", "Y", "N", "N", "...
## $ mkioskty2 <chr> "N", "N", "N", "N", "N", "N", "Y", "N", "N", "N", "N", "...
## $ mkioskty3 <chr> "Y", "Y", "N", "N", "N", "N", "N", "Y", "Y", "Y", "N", "...
## $ mkioskty4 <chr> "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "Y", "...
## $ mkioskty5 <chr> "Y", "Y", "N", "N", "N", "N", "N", "Y", "Y", "Y", "N", "...
## $ mkioskty6 <chr> "Y", "Y", "Y", "N", "N", "Y", "Y", "Y", "Y", "Y", "N", "...
## $ mkioskty7 <chr> "N", "Y", "N", "N", "N", "Y", "N", "Y", "N", "N", "N", "...
## $ mkioskty8 <chr> "Y", "Y", "N", "Y", "N", "Y", "N", "Y", "Y", "Y", "N", "...
## $ mkioskty9 <chr> "Y", "Y", "Y", "N", "N", "N", "N", "Y", "Y", "Y", "Y", "...
## $ mkioskty10 <chr> "N", "Y", "N", "N", "N", "N", "N", "Y", "Y", "N", "N", "...
## $ mkioskty11 <chr> "Y", "N", "N", "N", "Y", "N", "N", "Y", "Y", "N", "N", "...str(hpdata)## tibble [313 x 36] (S3: tbl_df/tbl/data.frame)
## $ dutyName.x: chr [1:313] "의료법인강릉동인병원" "강릉아산병원" "강원도삼척의료원" "강원도속초의료원" ...
## $ hpid : chr [1:313] "A2200005" "A2200008" "A2200007" "A2200012" ...
## $ hv10 : chr [1:313] "0" "Y" "0" "0" ...
## $ hv11 : chr [1:313] "0" "Y" "Y" "0" ...
## $ hv2 : num [1:313] 6 0 0 0 0 0 0 1 0 0 ...
## $ hv3 : num [1:313] 0 1 0 0 0 0 0 2 0 0 ...
## $ hv4 : num [1:313] 0 0 0 0 0 0 0 0 0 0 ...
## $ hv5 : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ hv6 : num [1:313] 0 1 0 0 0 0 0 1 1 0 ...
## $ hv7 : chr [1:313] "Y" "Y" "0" "0" ...
## $ hv8 : num [1:313] 0 0 0 0 0 0 0 0 0 0 ...
## $ hv9 : num [1:313] 0 0 0 0 0 0 0 4 0 0 ...
## $ hvamyn : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ hvangioayn: chr [1:313] "N" "N" "N" "N" ...
## $ hvcc : num [1:313] 0 0 0 0 0 0 0 0 0 0 ...
## $ hvccc : num [1:313] 0 0 0 0 0 0 0 0 0 0 ...
## $ hvctayn : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ hvec : num [1:313] 17 14 18 19 8 4 8 15 14 16 ...
## $ hvgc : num [1:313] 64 62 15 59 37 148 90 91 143 42 ...
## $ hvicc : num [1:313] 1 0 1 3 6 3 0 0 0 0 ...
## $ hvmriayn : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ hvncc : num [1:313] 0 6 0 0 0 0 0 17 10 0 ...
## $ hvoc : num [1:313] 5 13 2 3 4 3 6 15 11 9 ...
## $ hvventiayn: chr [1:313] "Y" "Y" "Y" "Y" ...
## $ mkioskty25: chr [1:313] "Y" "Y" "N" "Y" ...
## $ mkioskty1 : chr [1:313] "N" "Y" "Y" "N" ...
## $ mkioskty2 : chr [1:313] "N" "N" "N" "N" ...
## $ mkioskty3 : chr [1:313] "Y" "Y" "N" "N" ...
## $ mkioskty4 : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ mkioskty5 : chr [1:313] "Y" "Y" "N" "N" ...
## $ mkioskty6 : chr [1:313] "Y" "Y" "Y" "N" ...
## $ mkioskty7 : chr [1:313] "N" "Y" "N" "N" ...
## $ mkioskty8 : chr [1:313] "Y" "Y" "N" "Y" ...
## $ mkioskty9 : chr [1:313] "Y" "Y" "Y" "N" ...
## $ mkioskty10: chr [1:313] "N" "Y" "N" "N" ...
## $ mkioskty11: chr [1:313] "Y" "N" "N" "N" ...length of unique values in each variable
nuniq <- c()
for(i in 1:length(colnames(hpdata))) {
nuniq[i] <- hpdata[,i] %>%
n_distinct()
}
nuniq## [1] 312 313 3 3 13 12 1 2 9 3 1 10 1 1 5 6 2 35 140
## [20] 18 3 21 29 2 2 2 2 2 2 2 2 2 2 2 2 2remove columns with zero variance
hpdata <- hpdata[,nuniq!=1]str(hpdata)## tibble [313 x 32] (S3: tbl_df/tbl/data.frame)
## $ dutyName.x: chr [1:313] "의료법인강릉동인병원" "강릉아산병원" "강원도삼척의료원" "강원도속초의료원" ...
## $ hpid : chr [1:313] "A2200005" "A2200008" "A2200007" "A2200012" ...
## $ hv10 : chr [1:313] "0" "Y" "0" "0" ...
## $ hv11 : chr [1:313] "0" "Y" "Y" "0" ...
## $ hv2 : num [1:313] 6 0 0 0 0 0 0 1 0 0 ...
## $ hv3 : num [1:313] 0 1 0 0 0 0 0 2 0 0 ...
## $ hv5 : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ hv6 : num [1:313] 0 1 0 0 0 0 0 1 1 0 ...
## $ hv7 : chr [1:313] "Y" "Y" "0" "0" ...
## $ hv9 : num [1:313] 0 0 0 0 0 0 0 4 0 0 ...
## $ hvcc : num [1:313] 0 0 0 0 0 0 0 0 0 0 ...
## $ hvccc : num [1:313] 0 0 0 0 0 0 0 0 0 0 ...
## $ hvctayn : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ hvec : num [1:313] 17 14 18 19 8 4 8 15 14 16 ...
## $ hvgc : num [1:313] 64 62 15 59 37 148 90 91 143 42 ...
## $ hvicc : num [1:313] 1 0 1 3 6 3 0 0 0 0 ...
## $ hvmriayn : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ hvncc : num [1:313] 0 6 0 0 0 0 0 17 10 0 ...
## $ hvoc : num [1:313] 5 13 2 3 4 3 6 15 11 9 ...
## $ hvventiayn: chr [1:313] "Y" "Y" "Y" "Y" ...
## $ mkioskty25: chr [1:313] "Y" "Y" "N" "Y" ...
## $ mkioskty1 : chr [1:313] "N" "Y" "Y" "N" ...
## $ mkioskty2 : chr [1:313] "N" "N" "N" "N" ...
## $ mkioskty3 : chr [1:313] "Y" "Y" "N" "N" ...
## $ mkioskty4 : chr [1:313] "Y" "Y" "Y" "Y" ...
## $ mkioskty5 : chr [1:313] "Y" "Y" "N" "N" ...
## $ mkioskty6 : chr [1:313] "Y" "Y" "Y" "N" ...
## $ mkioskty7 : chr [1:313] "N" "Y" "N" "N" ...
## $ mkioskty8 : chr [1:313] "Y" "Y" "N" "Y" ...
## $ mkioskty9 : chr [1:313] "Y" "Y" "Y" "N" ...
## $ mkioskty10: chr [1:313] "N" "Y" "N" "N" ...
## $ mkioskty11: chr [1:313] "Y" "N" "N" "N" ...nuniq <- c()
for(i in 1:length(colnames(hpdata))) {
nuniq[i] <- hpdata[,i] %>%
n_distinct()
}
nuniq## [1] 312 313 3 3 13 12 2 9 3 10 5 6 2 35 140 18 3 21 29
## [20] 2 2 2 2 2 2 2 2 2 2 2 2 2devide table
hpdata_f <- hpdata[,nuniq<=3]
hpdata_n <- hpdata[,nuniq>3]recoding
hpdata_f <- hpdata_f %>%
mutate_all(funs(recode(., 'N1'=0L, '0'=0L, 'N'=0L, '1'=1L, 'Y'=1L, .default=1L)))## Warning: `funs()` is deprecated as of dplyr 0.8.0.
## Please use a list of either functions or lambdas:
##
## # Simple named list:
## list(mean = mean, median = median)
##
## # Auto named with `tibble::lst()`:
## tibble::lst(mean, median)
##
## # Using lambdas
## list(~ mean(., trim = .2), ~ median(., na.rm = TRUE))
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_warnings()` to see where this warning was generated.str(hpdata_f)## tibble [313 x 19] (S3: tbl_df/tbl/data.frame)
## $ hv10 : int [1:313] 0 1 0 0 0 0 0 1 1 1 ...
## $ hv11 : int [1:313] 0 1 1 0 0 0 0 1 1 1 ...
## $ hv5 : int [1:313] 1 1 1 1 1 1 1 1 1 1 ...
## $ hv7 : int [1:313] 1 1 0 0 0 0 0 1 1 1 ...
## $ hvctayn : int [1:313] 1 1 1 1 1 1 1 1 1 1 ...
## $ hvmriayn : int [1:313] 1 1 1 1 1 1 1 1 1 1 ...
## $ hvventiayn: int [1:313] 1 1 1 1 1 1 1 1 1 1 ...
## $ mkioskty25: int [1:313] 1 1 0 1 1 0 1 1 1 1 ...
## $ mkioskty1 : int [1:313] 0 1 1 0 0 0 0 1 1 0 ...
## $ mkioskty2 : int [1:313] 0 0 0 0 0 0 1 0 0 0 ...
## $ mkioskty3 : int [1:313] 1 1 0 0 0 0 0 1 1 1 ...
## $ mkioskty4 : int [1:313] 1 1 1 1 1 1 1 1 1 1 ...
## $ mkioskty5 : int [1:313] 1 1 0 0 0 0 0 1 1 1 ...
## $ mkioskty6 : int [1:313] 1 1 1 0 0 1 1 1 1 1 ...
## $ mkioskty7 : int [1:313] 0 1 0 0 0 1 0 1 0 0 ...
## $ mkioskty8 : int [1:313] 1 1 0 1 0 1 0 1 1 1 ...
## $ mkioskty9 : int [1:313] 1 1 1 0 0 0 0 1 1 1 ...
## $ mkioskty10: int [1:313] 0 1 0 0 0 0 0 1 1 0 ...
## $ mkioskty11: int [1:313] 1 0 0 0 1 0 0 1 1 0 ...merge again
hpdata <- bind_cols(hpdata_n, hpdata_f)
glimpse(hpdata)## Rows: 313
## Columns: 32
## $ dutyName.x <chr> "의료법인강릉동인병원", "강릉아산병원", "강원도삼척의료원", "강원도속초의료원", "의료법인보광의...
## $ hpid <chr> "A2200005", "A2200008", "A2200007", "A2200012", "A220004...
## $ hv2 <dbl> 6, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 3, 0, 0,...
## $ hv3 <dbl> 0, 1, 0, 0, 0, 0, 0, 2, 0, 0, 0, 0, 0, 0, 5, 10, 4, 0, 0...
## $ hv6 <dbl> 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hv9 <dbl> 0, 0, 0, 0, 0, 0, 0, 4, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hvcc <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hvccc <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
## $ hvec <dbl> 17, 14, 18, 19, 8, 4, 8, 15, 14, 16, 9, 10, 10, 10, 13, ...
## $ hvgc <dbl> 64, 62, 15, 59, 37, 148, 90, 91, 143, 42, 73, 19, 30, 10...
## $ hvicc <dbl> 1, 0, 1, 3, 6, 3, 0, 0, 0, 0, 11, 1, 0, 6, 0, 0, 0, 4, 4...
## $ hvncc <dbl> 0, 6, 0, 0, 0, 0, 0, 17, 10, 0, 0, 0, 0, 3, 13, 4, -1, 0...
## $ hvoc <dbl> 5, 13, 2, 3, 4, 3, 6, 15, 11, 9, 2, 2, 0, 8, 10, 2, 13, ...
## $ hv10 <int> 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 0,...
## $ hv11 <int> 0, 1, 1, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 0,...
## $ hv5 <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,...
## $ hv7 <int> 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 1,...
## $ hvctayn <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,...
## $ hvmriayn <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,...
## $ hvventiayn <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,...
## $ mkioskty25 <int> 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 0,...
## $ mkioskty1 <int> 0, 1, 1, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0,...
## $ mkioskty2 <int> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0,...
## $ mkioskty3 <int> 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 0, 0, 0,...
## $ mkioskty4 <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,...
## $ mkioskty5 <int> 1, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 1, 0, 1, 1, 0,...
## $ mkioskty6 <int> 1, 1, 1, 0, 0, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 0, 0, 1, 0,...
## $ mkioskty7 <int> 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0,...
## $ mkioskty8 <int> 1, 1, 0, 1, 0, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 0,...
## $ mkioskty9 <int> 1, 1, 1, 0, 0, 0, 0, 1, 1, 1, 1, 1, 0, 1, 1, 0, 0, 1, 0,...
## $ mkioskty10 <int> 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0,...
## $ mkioskty11 <int> 1, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0,...centering and scaling
hpdata_z <- hpdata %>%
mutate_each_(funs(scale), vars=colnames(hpdata)[3:32])## Warning: `mutate_each_()` is deprecated as of dplyr 0.7.0.
## Please use `across()` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_warnings()` to see where this warning was generated.head(hpdata_z[,3:32])## # A tibble: 6 x 30
## hv2[,1] hv3[,1] hv6[,1] hv9[,1] hvcc[,1] hvccc[,1] hvec[,1] hvgc[,1] hvicc[,1]
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 3.12 -0.356 -0.235 -0.171 -0.149 -0.124 1.14 -0.364 -0.490
## 2 -0.359 0.120 0.642 -0.171 -0.149 -0.124 0.654 -0.397 -0.762
## 3 -0.359 -0.356 -0.235 -0.171 -0.149 -0.124 1.30 -1.19 -0.490
## 4 -0.359 -0.356 -0.235 -0.171 -0.149 -0.124 1.46 -0.448 0.0548
## 5 -0.359 -0.356 -0.235 -0.171 -0.149 -0.124 -0.318 -0.819 0.871
## 6 -0.359 -0.356 -0.235 -0.171 -0.149 -0.124 -0.967 1.05 0.0548
## # ... with 21 more variables: hvncc[,1] <dbl>, hvoc[,1] <dbl>, hv10[,1] <dbl>,
## # hv11[,1] <dbl>, hv5[,1] <dbl>, hv7[,1] <dbl>, hvctayn[,1] <dbl>,
## # hvmriayn[,1] <dbl>, hvventiayn[,1] <dbl>, mkioskty25[,1] <dbl>,
## # mkioskty1[,1] <dbl>, mkioskty2[,1] <dbl>, mkioskty3[,1] <dbl>,
## # mkioskty4[,1] <dbl>, mkioskty5[,1] <dbl>, mkioskty6[,1] <dbl>,
## # mkioskty7[,1] <dbl>, mkioskty8[,1] <dbl>, mkioskty9[,1] <dbl>,
## # mkioskty10[,1] <dbl>, mkioskty11[,1] <dbl>2. PCA
Principal Component Analysis
hp_without_id <- hpdata_z[,3:32] %>%
as.matrix()
hp_pca <- prcomp(hp_without_id)
hp_pca[[1]]## [1] 2.754002e+00 1.720755e+00 1.411599e+00 1.242049e+00 1.144886e+00
## [6] 1.128249e+00 1.069838e+00 1.037569e+00 1.009868e+00 9.689509e-01
## [11] 9.242822e-01 9.118337e-01 8.872557e-01 8.452841e-01 8.235437e-01
## [16] 8.171788e-01 7.921716e-01 7.759205e-01 6.981109e-01 6.681701e-01
## [21] 6.511012e-01 6.362573e-01 5.868942e-01 5.730135e-01 5.228457e-01
## [26] 5.158809e-01 3.967430e-01 3.825128e-01 3.376772e-01 1.223023e-15hp_pca[[2]][,1:3]## PC1 PC2 PC3
## hv2 -0.18428779 0.13822699 -0.060154202
## hv3 -0.20384875 0.19991856 -0.086350699
## hv6 -0.17148933 0.14451215 -0.039068239
## hv9 -0.09808548 0.01830479 0.010927088
## hvcc -0.09101464 0.14558902 -0.062165691
## hvccc -0.12159677 0.10212124 -0.068971427
## hvec -0.01194454 -0.23947804 0.036179058
## hvgc -0.17597517 0.09361233 -0.139507068
## hvicc 0.04669590 -0.24742423 0.006235784
## hvncc -0.26006542 0.15943534 -0.022511889
## hvoc -0.27206005 0.10386183 -0.027060730
## hv10 -0.30026561 0.14313398 -0.006508880
## hv11 -0.28773147 0.13032689 -0.013353870
## hv5 -0.03921070 -0.18245036 -0.649749438
## hv7 -0.24181735 -0.12239585 0.050785418
## hvctayn -0.03921070 -0.18245036 -0.649749438
## hvmriayn -0.10320804 -0.20778141 -0.171891569
## hvventiayn -0.01475009 0.01723712 0.012073206
## mkioskty25 -0.14504399 -0.32545834 0.093917439
## mkioskty1 -0.30002855 0.09324527 0.035885149
## mkioskty2 -0.01240845 -0.17689169 0.072179790
## mkioskty3 -0.20985983 -0.27106174 0.126248272
## mkioskty4 -0.03275784 -0.08519503 0.009874346
## mkioskty5 -0.23969715 -0.20245868 0.115246037
## mkioskty6 -0.11951641 -0.33596619 0.099173657
## mkioskty7 -0.12034658 -0.09620336 0.051132989
## mkioskty8 -0.14749307 -0.29630323 0.082464088
## mkioskty9 -0.17809874 -0.26653048 0.117523007
## mkioskty10 -0.29958953 0.10095414 0.031058263
## mkioskty11 -0.23728115 0.05708709 0.033242411variance explained: R2
First conponent explains 25% of the total variance.
summary(hp_pca)## Importance of components:
## PC1 PC2 PC3 PC4 PC5 PC6 PC7
## Standard deviation 2.7540 1.7208 1.41160 1.24205 1.14489 1.12825 1.06984
## Proportion of Variance 0.2528 0.0987 0.06642 0.05142 0.04369 0.04243 0.03815
## Cumulative Proportion 0.2528 0.3515 0.41794 0.46936 0.51305 0.55548 0.59364
## PC8 PC9 PC10 PC11 PC12 PC13 PC14
## Standard deviation 1.03757 1.00987 0.9690 0.92428 0.91183 0.88726 0.84528
## Proportion of Variance 0.03588 0.03399 0.0313 0.02848 0.02771 0.02624 0.02382
## Cumulative Proportion 0.62952 0.66352 0.6948 0.72329 0.75100 0.77724 0.80106
## PC15 PC16 PC17 PC18 PC19 PC20 PC21
## Standard deviation 0.82354 0.81718 0.79217 0.77592 0.69811 0.66817 0.65110
## Proportion of Variance 0.02261 0.02226 0.02092 0.02007 0.01625 0.01488 0.01413
## Cumulative Proportion 0.82367 0.84593 0.86684 0.88691 0.90316 0.91804 0.93217
## PC22 PC23 PC24 PC25 PC26 PC27 PC28
## Standard deviation 0.63626 0.58689 0.57301 0.52285 0.51588 0.39674 0.38251
## Proportion of Variance 0.01349 0.01148 0.01094 0.00911 0.00887 0.00525 0.00488
## Cumulative Proportion 0.94567 0.95715 0.96809 0.97720 0.98608 0.99132 0.99620
## PC29 PC30
## Standard deviation 0.3377 1.223e-15
## Proportion of Variance 0.0038 0.000e+00
## Cumulative Proportion 1.0000 1.000e+00scree plot
screeplot(hp_pca, main = "", col = "blue", type = "lines", pch = 21)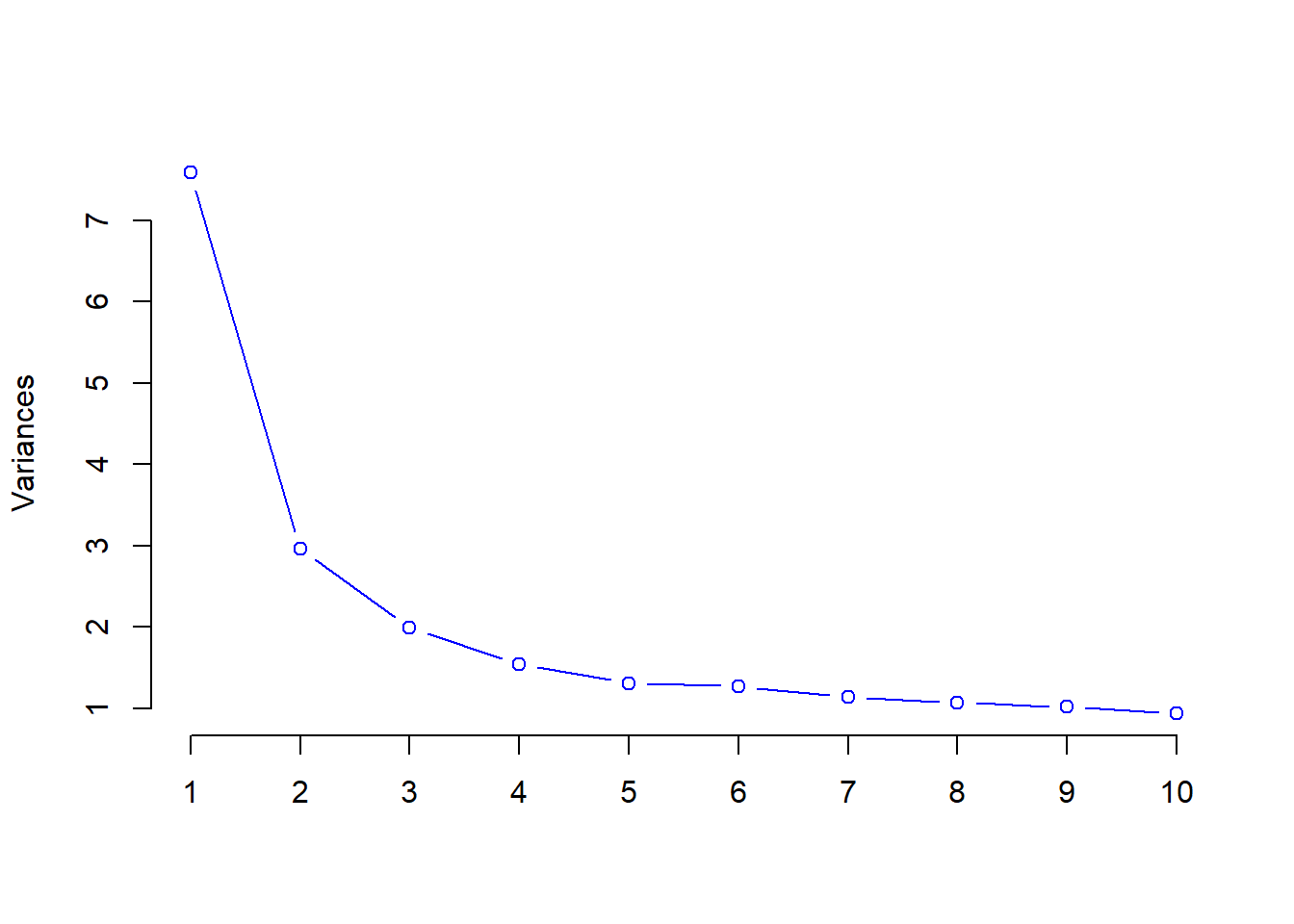
hospital score
hp_pc1 <- predict(hp_pca)[,1]
hp_score <- (100-20*hp_pc1)
hospital_score <- hpdata %>%
select(dutyName.x,hpid)%>%
mutate(score=hp_score)
skim(hospital_score)| Name | hospital_score |
| Number of rows | 313 |
| Number of columns | 3 |
| _______________________ | |
| Column type frequency: | |
| character | 2 |
| numeric | 1 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| dutyName.x | 0 | 1 | 3 | 23 | 0 | 312 | 0 |
| hpid | 0 | 1 | 8 | 8 | 0 | 313 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| score | 0 | 1 | 100 | 55.08 | -4.89 | 56.88 | 82.43 | 127.78 | 301.22 | ▅▇▂▂▁ |
plotting hospital scores
library(ggplot2)
plot_score <- ggplot(hospital_score, aes(x=score))+
geom_histogram(fill='sky blue', binwidth = 10)
plot_score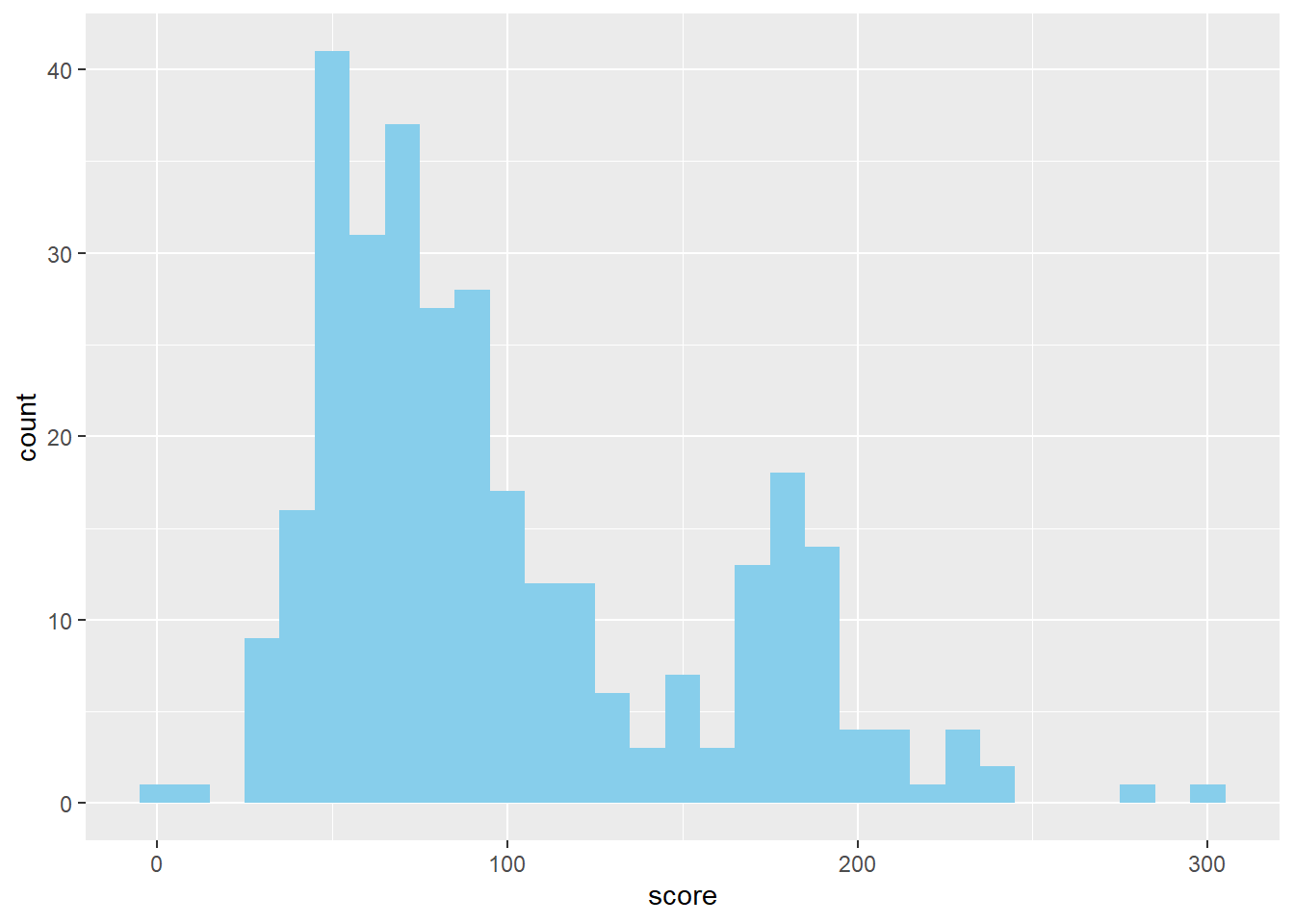
exporting dataset
write.csv(hospital_score, file = 'hospital_score')